Quick Guide Contents
Introduction
The Super Pan-genome of Wild & Cultivated Rice Project based on reference-quality de novo long-read assemblies of 251 rice accessions (202 O. sativa, 28 O. rufipogon, 11 O. glaberrima, and 10 O. barthii) aimed to generate a comprehensive and complete super pan-genome, super pan-annotation and super pan-structural-variations (super pan-SVs). To facilitate the applications of these results, we developed the Rice Super Pan-genome Information Resource Database, an interactive web-based browser for the rice super pan-genome. The reference-free whole-genomes alignments of 251 accessions, genome annotation, structure variations, pan-genome graph and K-mer spectrums were also included in the RiceSuperPIRdb. Besides, more data and analysis results will also be included in our database soon.
User Manual
The RiceSuperPIRdb provides the following data:
- 1. Basic information of the 251 rice accessions, including the genome sequence, gene annotation, and structural variation;
- 2. Reference-free multiple assembly alignment, which means that the user can choose any accession as a reference to browse whole genome alignment;
- 3. Pan-genome and annotation information, including a mainly multi-allelic pan-genome graph of rice material through minigraph and a mainly bi-allelic pan-genome graph through varation graph toolkit. Both pan-genomes have been annotated;
- 4. Node-specific k-mer spectrums for each of 256 accessions, including our 251 rice accessions and 5 public accessions (O.punctata, O.meridionalis, O.glumaepatula, Nipponbare, and R498), which can be used to verify their closest assembly reference with low-quality short-read data.
The RiceSuperPIRdb also provides the following functions:
- 1. Visualization function:
A Super Pan Browser visualizes the genome sequence, the gene annotation and the multi genome sequence alignment results. It allows users to select any accession as a reference to view the whole genome alignment result. As the analysis process is a time-consuming process, in order to save the users’ time, the whole-genomes alignment result based on Nipponbare or R498 has been already prepared well. The user can also view the pan-genome annotation and the structural variation results while selecting the Nipponbare reference. - 2. Download function:
Search a single rice accession (with its accession code) to obtain its genome sequence, gene annotation and SV information. The user can also download the meta-information of k-mer spectrums and pan-genome information. - 3. Blast function.
There are the basic search and advanced search providing to the users.
SUPER PAN BROWSER
Three selections, one of 251 accessions, Nipponbare and R498 were provided on the Rice Hub Assembly part.
1.Selecting one of the 251 assemblies as a reference.
When the users select one of the 251 assemblies, the analyze result would be transformed online, which could be a time-consuming process, so we provided two ways, interactive or sending an e-mail for the users to choose.
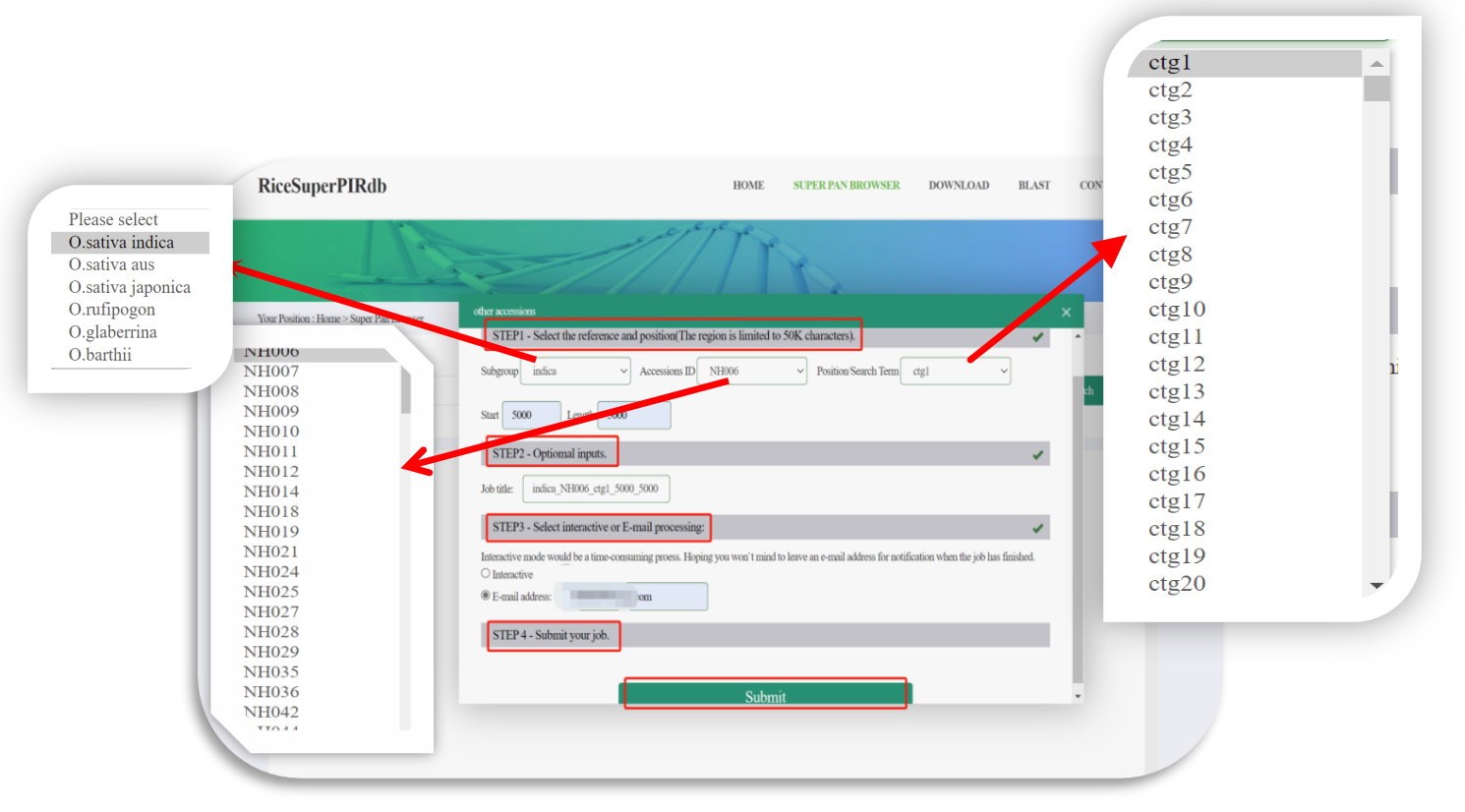
Figure 1. Example of selecting one of 251 assemblies.
Then you will receive an e-mail after a few minutes or several hours, since it depends on the length which the users would like to view.
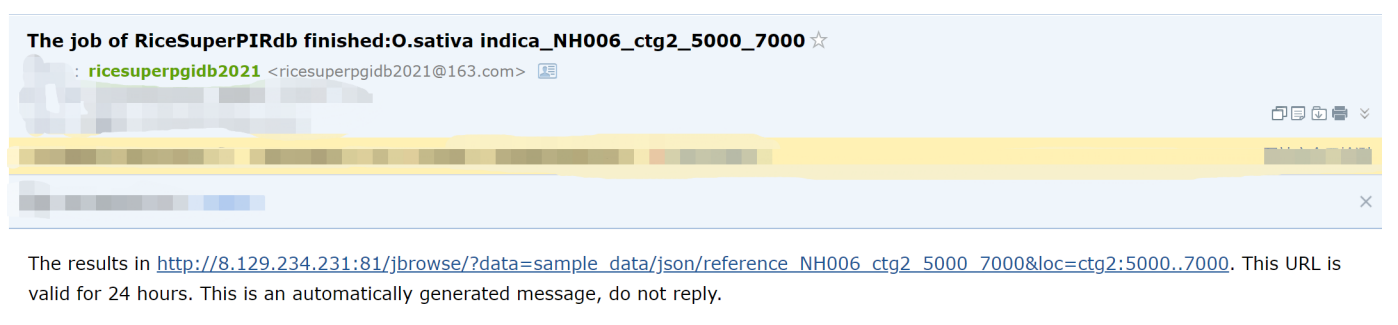
Figure 2. Example of E-mail received by the users.
2.Selecting the Nipponbare or R498 as a reference.
If the Nipponbare was selected, the multi-genome alignment, pan-genome annotation, SVs information, and the genome sequence would be illustrated on the web. Considering the assess speed and best performance, we recommend the users to select the length less than 4000bp each time.
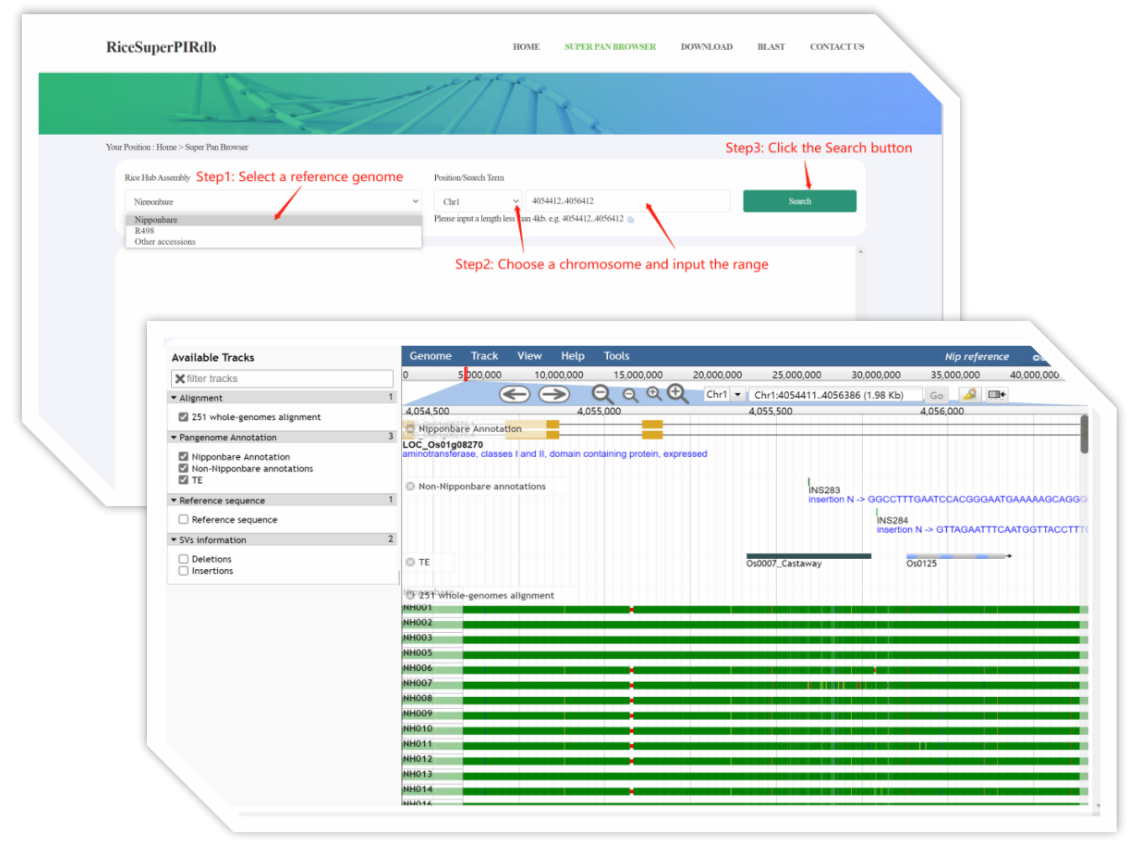
Figure 3. Example of using the SUPER PAN BROWSER channel.
(1)Some basic use of the Jbrowse.
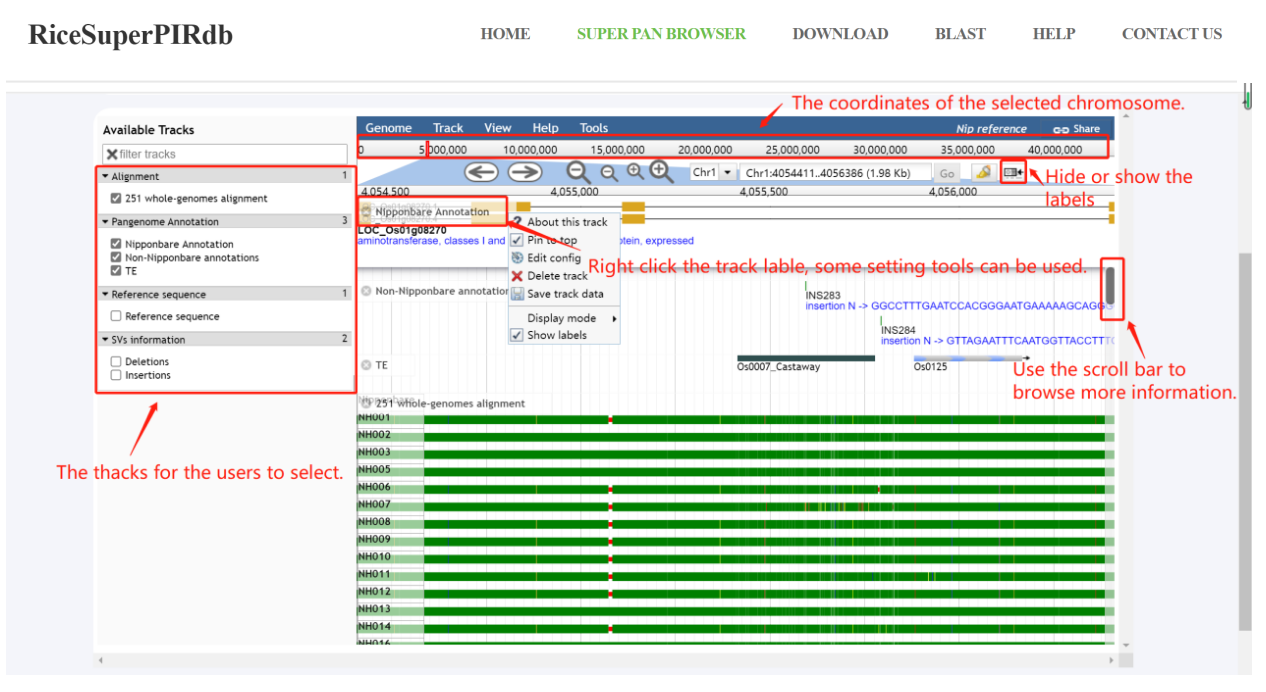
Figure 4. A introducing sample of using the Jbrowse.
(2)The introduction of the Alignment track.
The blank spaces are deletion variations in some rice genomes compared with Nipponbare. The blue, red, orange, and yellow showing in the alignment graph instead of the mismatch of A, C, G, and T respectively.
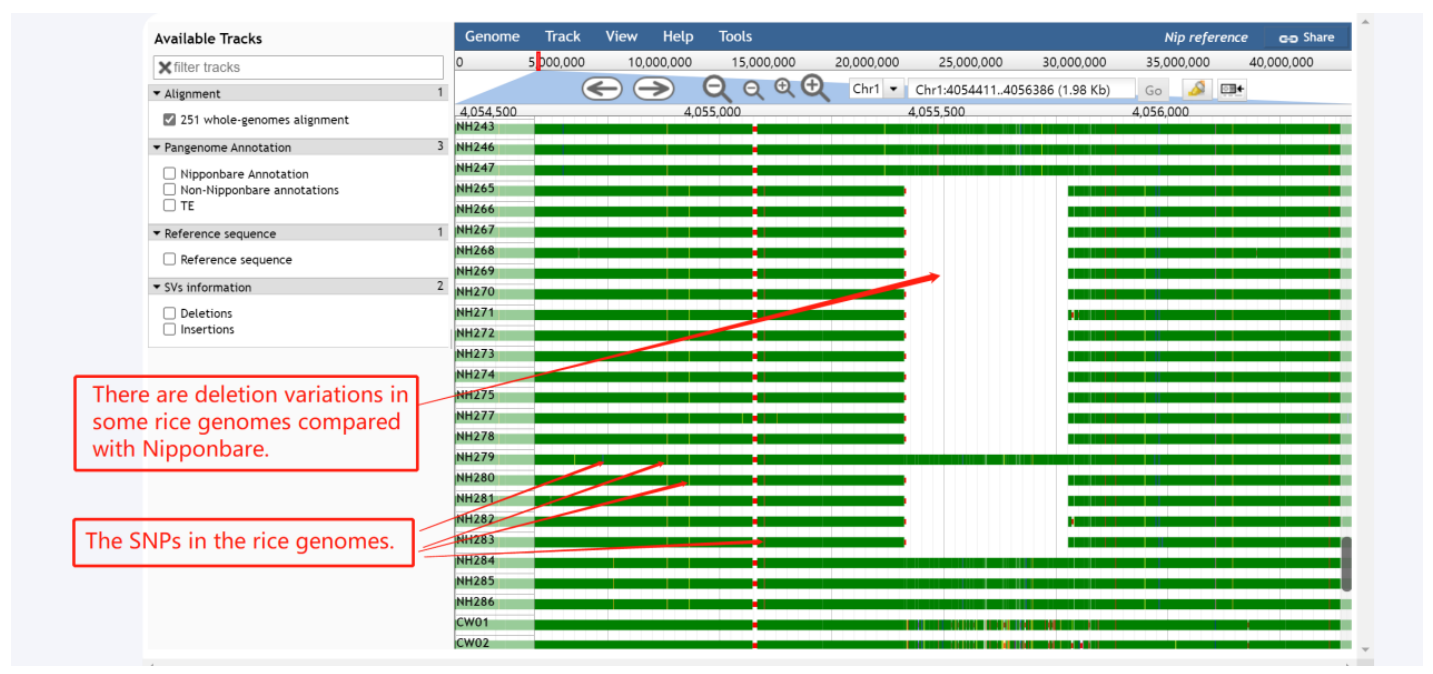
Figure 5. Example of using the Alignment track of SUPER PAN BROWSER channel.
(3)The introduction of the Pan-genome Annotation.
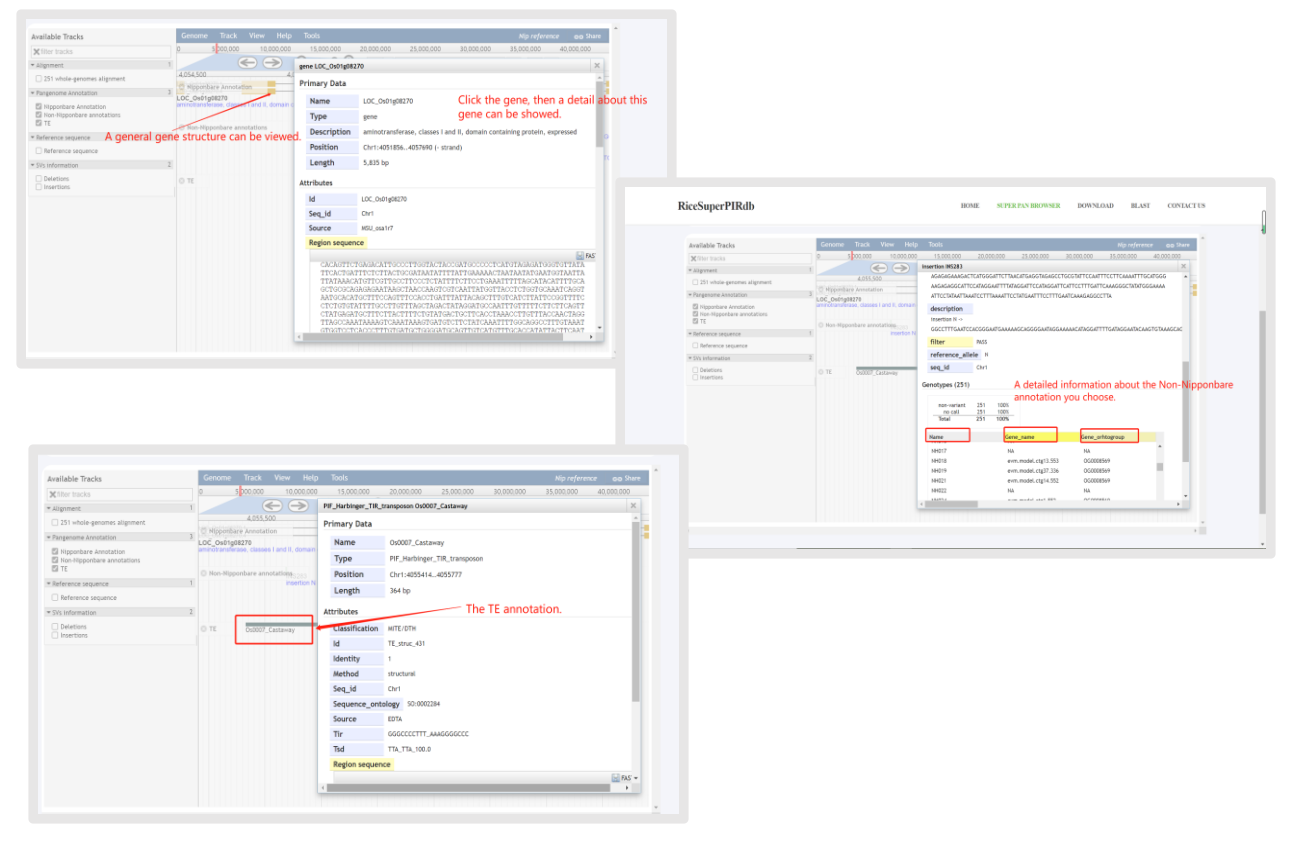
Figure 6. Example of using the Pan-genome Annotation track of SUPER PAN BROWSER channel.
(4)The SVs information track.
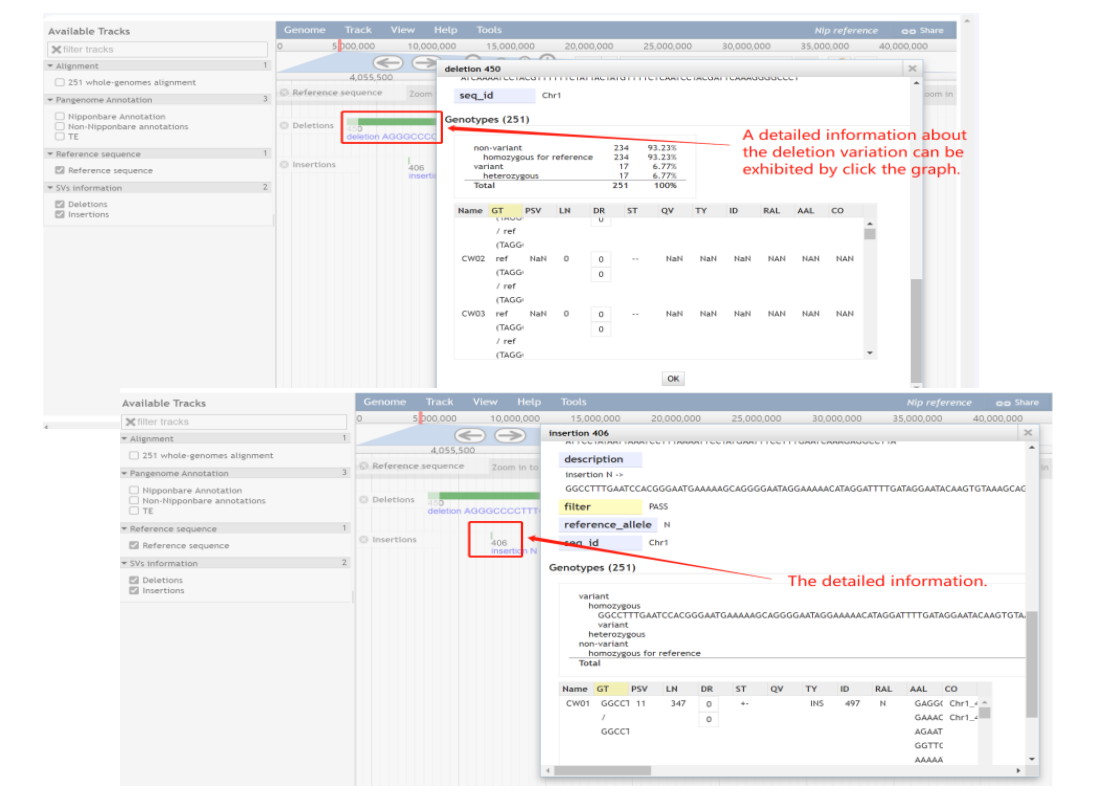
Figure 7. Example of using the SVs information track of SUPER PAN BROWSER channel.
DOWNLOAD
The sequences, gene annotations, and structural variation of the 251 individual, the K-mer spectrums, as well as the pan-genome and annotation are available on the download page.
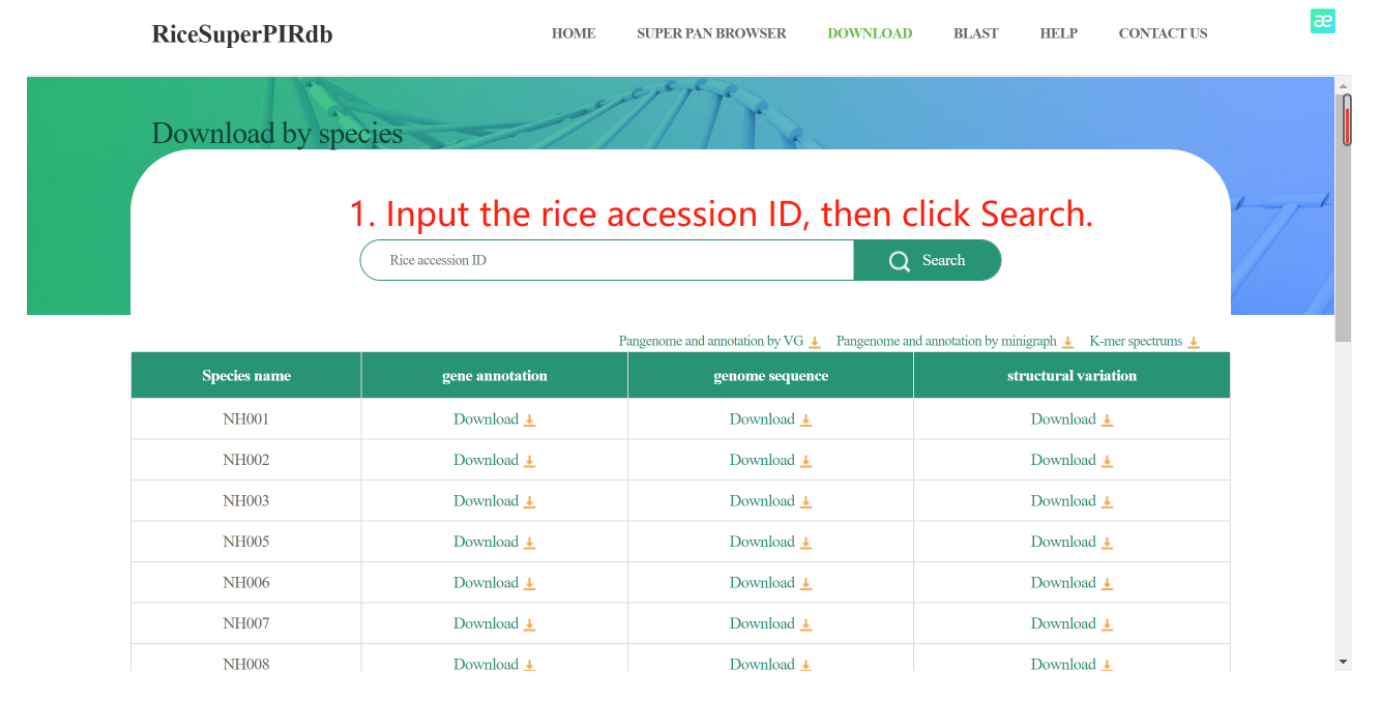
Figure 8. The usage of the DOWNLOAD page.
BLAST
BLAST finds regions of similarity between biological sequences. The program compares nucleotide or protein sequences to sequence databases (the 251 cultivated and wild Asian and African rice accessions) and calculates the statistical significance.
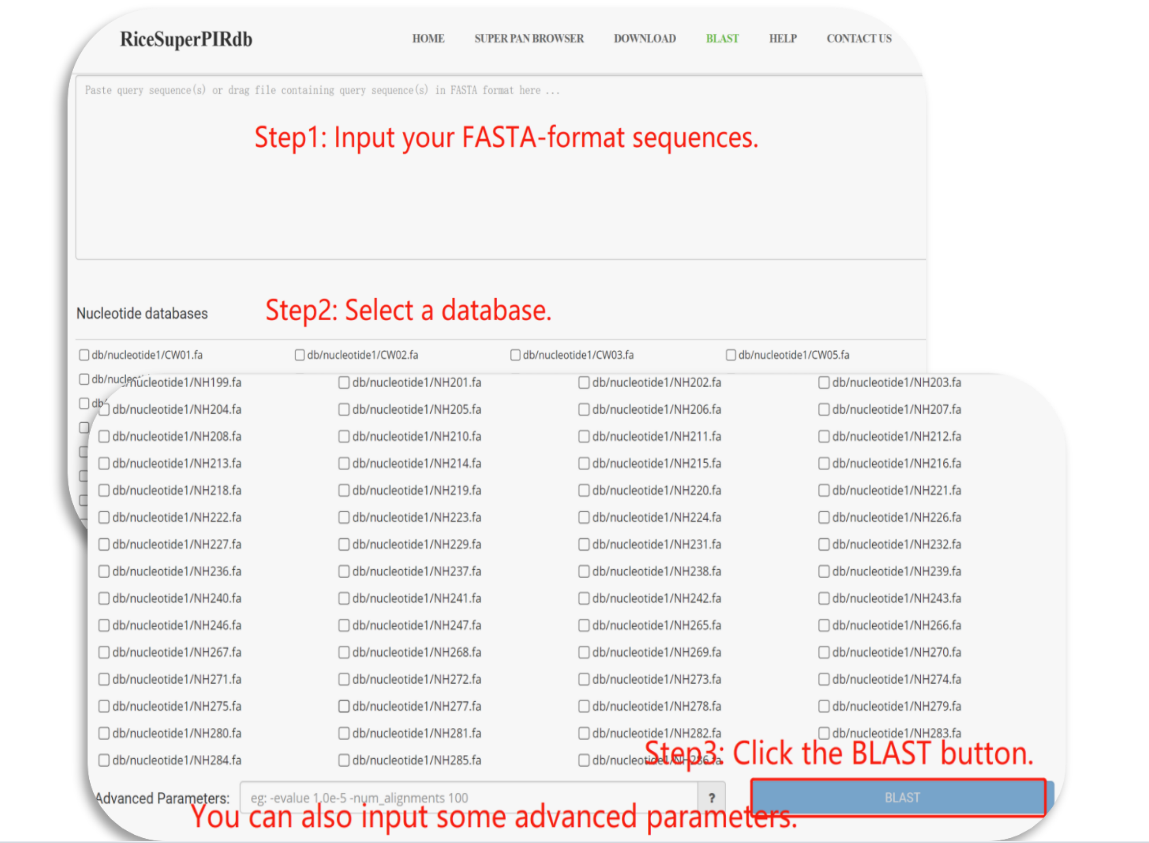
Figure 8. The introduction of the BLAST tool.
The blast results are as follows:
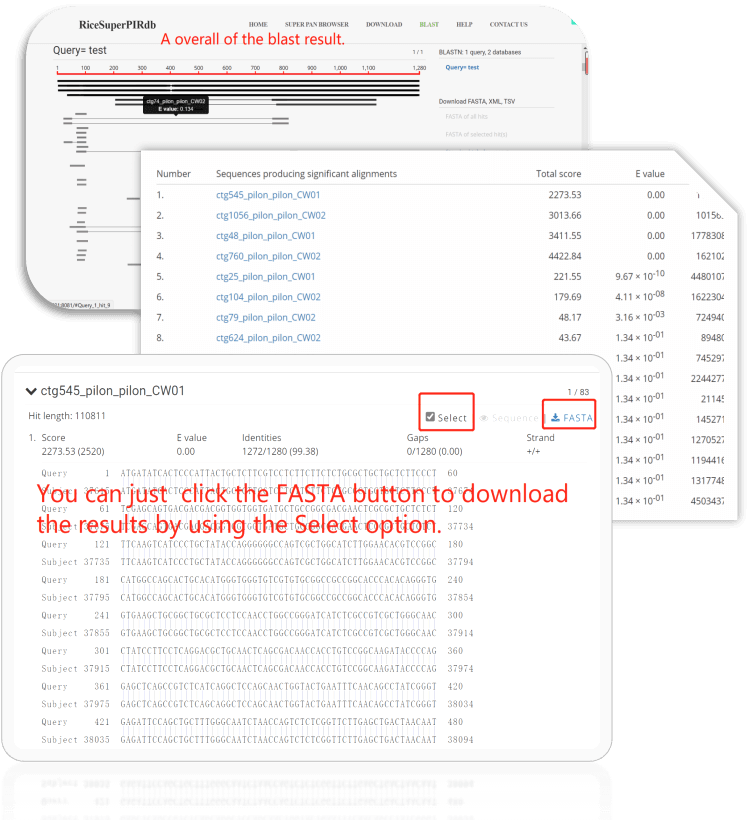
Figure 9. Example of using the BLAST tool.
Database update
- 2021.7.9
Construct the Pan-genome and annotation browser.